Bacteria Testing: Genetic Methods
The first line of defense against microbial proliferation is the measurement of actively growing microorganisms. The medical and food industry have used genetic methods as a means of testing for years to obtain results more efficiently and effectively than culture methods; however, this has not been the case with the oil industry.
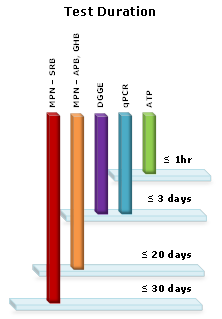
NACE TM0194 is the current standard followed by the industry for measuring bacteria in the system. This standard uses the Most Probable Number (MPN) method to quantify the bacteria in the sample. Typically, only a small percentage of the actual bacteria population grows in culture media in a laboratory and the test only quantifies what can grow in the laboratory media instead of what is actually in the sample. Results are obtained 14 to 28 days after the samples are inoculated, a major disadvantage if there is a real problem in the system.
The Problem With Bacteria
Bacteria are contained in all oilfield water systems. The three major problems that result from the growth of bacteria are microbiologically induced corrosion (MIC), reservoir souring and fouling. All are the result of complex phenomena comprising of a range of mechanisms and microbe types and interrelationships. Common groups of bacteria that are of major importance to the oilfield include Sulfate Reducing Bacteria (SRB), Heterotrophic Aerobic Bacteria (HAB), Iron Reducing and Iron Oxidizing Bacteria (IRB/IOB), Slime Formers, Acid Producing Bacteria (APB), General Heterotrophic Bacteria (GHB), and Nitrate Reducing Bacteria (NRB).
Common mechanisms that promote MIC include hydrogen sulfide (H2S) generation, lowering of pH, breakdown of passivating oxide films, under-deposit corrosion, differential aeration, and direct oxidation of metallic substrates.
Sulfate-Reducing Bacteria (SRB) contribute to MIC through the production of H2S. H2S is extremely corrosive, especially in gas systems. Focusing on SRB detection at the exclusion of other bacteria is understandable due to their activity and ease of detection, but to ignore the non-SRB members of the microbial community only describes a partial picture of the bacteria present. Sulfide is not only produced by SRB, but also by a group of Archaea, methanogens, and even fermentative microorganisms in the system.
Other H2S Generators
Sulfide is not only produced by SRB, but also by a group of Archaea, methanogens, and even fermentative microorganisms in the system. The proportion of H2S generated by sulfate reducing Archaea (SRA) relative to SRB is currently unknown. Also, the proportion of H2S generated by fermenters and methanogens relative to H2S produced by SRB is unknown. However, it has become clear that H2S is generated both in oil field reservoirs and in topside facilities by a much more diverse community of prokaryotes than previously acknowledged by the oil industry.
How Genetic Tests Can Help
While the MPN method quantifies what can grow in laboratory media, genetic tests quantify what is actually in the sample, giving a more reliable picture of the system at its current state. Entire populations, most abundant species, or specific bacteria can be identified with genetic testing. Plus, some types of organisms that contribute to MIC can’t be conveniently quantified using MPN tests, but genetic tests can achieve this. Although the upfront costs of genetic testing can be high, they may be less when personnel time and materials are factored in.
While genetic tests are able to give quick answers on the microbiological community, not all genetic methods are able to be performed offshore. Highly-trained staff are needed to perform testing. Hence, samples will need to be fixed on site and shipped to the laboratory for analysis in a timely manner to obtain reliable results.
Adenosine Triphosphate (ATP) Photometry
Adenosine Triphosphate (ATP) Photometry allows the rapid determination of all living organisms present in a water sample. This test is not specific to bacteria, but since a majority of living organisms in the oilfield are bacteria, it can be a useful technique. ATP is a direct test, meaning that it avoids the growing of organisms in “captivity” and tests what is actually in the sample. Since the test does not identify the type of bacteria present, other testing methods need to be used in conjunction with ATP. The test takes as little as 30 minutes to complete and can be performed in the field provided that there are trained personnel to perform the test.
ATP Photometry measures the amount of ATP present in the sample. During the test, luciferin and luciferase are introduced into the sample which reacts with the ATP in the presence of oxygen to produce a bioluminescence. The amount of light emitted is measured with a photometer and the number of organisms determined from calibration data.
ATP results indicate the relative abundance of metabolically-active total bacteria. A large number of dormant bacteria will yield a low ATP concentration while a smaller number of metabolically active bacteria will yield a higher ATP concentration. There is no direct conversion of ATP concentrations to microbial concentrations. Moreover, ATP values reflect the activity of the entire microbial population and cannot distinguish what amount of activity is present in certain types of bacteria, such as SRB.
Quantitative Polymerase Chain Reaction (qPCR)
Quantitative Polymerase Chain Reaction (qPCR) amplifies specific gene sequences from target organisms. During qPCR, genetic material is extracted from a sample and subsequently the numbers of copies of a specific gene in the extract are quantified. Thus, unlike all the other methods, qPCR does not rely on visualization, viability, or separation of individual organisms. qPCR is also well suited for difficult samples, including solids, corrosion products or produced water. qPCR delivers the number of total bacteria, total Archaea, SRB and SRA, as well as specific species under each group. qPCR does not distinguish between live, inactive and dead cells, making it hard to imply activity. Using qPCR along with ATP or MPN is recommended.
Denaturing Gradient Gel Electrophoresis (DGGE)
Denaturing Gradient Gel Electrophoresis (DGGE) is a DNA-based technique which generates a genetic profile or fingerprint of the microbial community. Individual sequences or bands from this profile can be extracted and sequenced to identify the dominant members of the microbial population. DGGE fingerprints can be produced and dominant microorganisms can be identified for a variety of target groups including bacteria, fungi, SRB and Dehalococcoides.
Uses of DGGE include being able to assess the changes in the population and bacteria types during biocide treatments. For example, if a biocide treatment is sent to kill a certain type of bacteria, a DGGE test can show what bacteria are left, if new bands have gained a foothold in the damaged biofilm, etc. If nitrate is injected, one may see several bands disappear, probably representing SRB, and new prominent bands appear, probably representing NRB.
Generally a bacterial species must be present at 1% or more of the total population in order to produce a band that is clearly visible on a DGGE gel. Therefore, the DGGE bands allow for the detection of the most abundant species of bacteria present in a sample and the intensity of the DGGE band is proportional to the relative abundance of the bacterial species in that sample.
Conclusion
Genetic techniques have demonstrated that we know little about the majority of the bacteria in our systems. NACE TM0194 may be updated with genetic testing methods in the near future. Over time, genetic testing can prove to be a cost-effective means of managing production and water injection systems. ATP testing offers reliable and quick determination of bacteria activity in the field without the need to wait 14 to 28 days for MPN test results.
While one test cannot give comprehensive results, a combination of testing including genetic testing will be able to provide a more reliable and complete picture of the bacteria in the system than the use of MPN alone.
References
Microbes in the Marine Industry. Edited by EC Hill. Copyright 2003.
J. Larsen; K.B. Sørensen; B. Højris; T.L. Skovhus, Significance of Troublesome Sulfate-Reducing Prokaryotes (SRP) in Oil Field Systems, Corrosion 2009, paper 09389, (Houston, TX: NACE International, 2009).
DGGE: Profile and Identify Dominant Members Within A Microbial Community. Accessed September 27, 2012. http://www.microbe.com/index.php/DGGE/dgge-er-overview.html.
Whitby, Corinne; Skovhus, Torben Lund. Applied Microbiology and Molecular Biology in Oilfield Systems. Springer Science+Business Media. January 1, 2011.